Our recent work in biomedical applications
Modeling Retinal Ganglion Cells with Neural Differential Equations
Kacper Dobek, Daniel Jankowski, and Krzysztof Krawiec
Accepted at Student Workshop at AAAI-26.
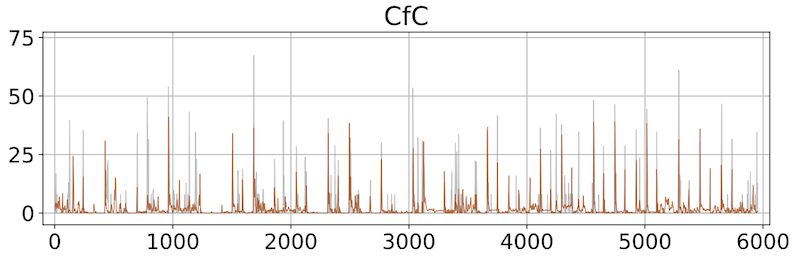
This work explores Liquid Time-Constant Networks (LTCs) and Closed-form Continuous-time Networks (CfCs) for modeling retinal ganglion cell activity in tiger salamanders across three datasets. Compared to a convolutional baseline and an LSTM, both architectures achieved lower MAE, faster convergence, smaller model sizes, and favorable query times, though with slightly lower Pearson correlation. Their efficiency and adaptability make them well suited for scenarios with limited data and frequent retraining, such as edge deployments in vision prosthetics.
Evidence Generation for a Host-Response Biosignature of Respiratory Disease
Kelly E. Dooley, Michael Morimoto, Piotr Kaszuba, Margaret Krasne, Gigi Liu, Edward Fuchs, Peter Rexelius, Jerry Swan, Krzysztof Krawiec, Kevin Hammond, Stuart C. Ray, Ryan Hafen, Andreas Schuh, and Nelson L. Shasha Jumbe
Viruses. 2025 Jul 2;17(7):943. https://pubmed.ncbi.nlm.nih.gov/40733560/
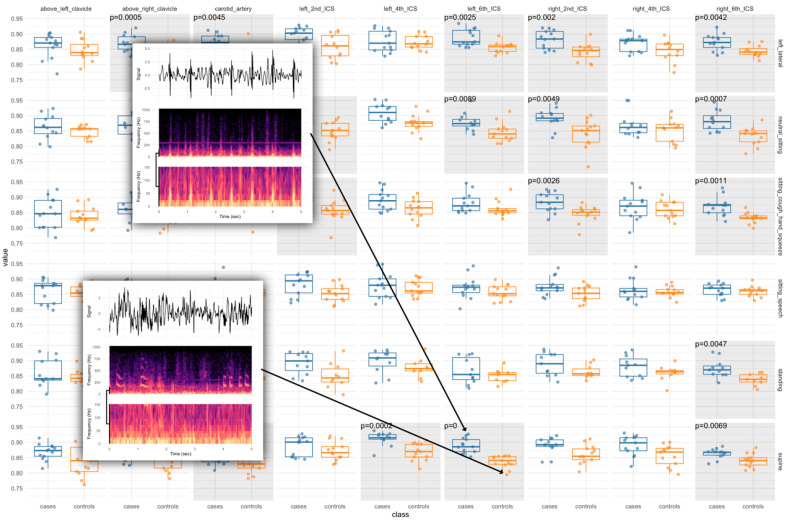
Background: In just twenty years, three dangerous human coronaviruses-SARS-CoV, MERS-CoV, and SARS-CoV-2 have exposed critical gaps in early detection of emerging viral threats. Current diagnostics remain pathogen-focused, often missing the earliest phase of infection. A virus-agnostic, host-based diagnostic capable of detecting responses to viral intrusion is urgently needed. Methods: We hypothesized that the lungs act as biomechanical instruments, with infection altering tissue tension, wave propagation, and flow dynamics in ways detectable through subaudible vibroacoustic signals. In a matched case-control study, we enrolled 19 RT-PCR-confirmed COVID-19 inpatients and 16 matched controls across two Johns Hopkins hospitals. Multimodal data were collected, including passive vibroacoustic auscultation, lung ultrasound, peak expiratory flow, and laboratory markers. Machine learning models were trained to identify host-response biosignatures from anterior chest recordings. Results: 19 COVID-19 inpatients and 16 matched controls (mean BMI 32.4 kg/m2, mean age 48.6 years) were successfully enrolled to the study. The top-performing, unoptimized, vibroacoustic-only model achieved an AUC of 0.84 (95% CI: 0.67-0.92). The host-covariate optimized model achieved an AUC of 1.0 (95% CI: 0.94-1.0), with 100% sensitivity (95% CI: 82-100%) and 99.6% specificity (95% CI: 85-100%). Vibroacoustic data from the anterior chest alone reliably distinguished COVID-19 cases from controls. Conclusions: This proof-of-concept study demonstrates that passive, noninvasive vibroacoustic biosignatures can detect host response to viral infection in a hospitalized population and supports further testing of this modality in broader populations. These findings support the development of scalable, host-based diagnostics to enable early, agnostic detection of future pandemic threats (ClinicalTrials.gov number: NCT04556149).
Synthesizing Effective Diagnostic Models from Small Samples Using Structural Machine Learning: A Case Study in Automating COVID-19 Diagnosis
Piotr Kaszuba, Andrew Turner, Bartosz Mikulski, Nl Shasha Jumbe, Andreas Schuh, Michael Morimoto, Peter Rexelius, Ryan Hafen, Ron Deiotte, Kevin Hammond, Jerry Swan, and Krzysztof Krawiec
GECCO '23, https://dl.acm.org/doi/10.1145/3583133.3590598
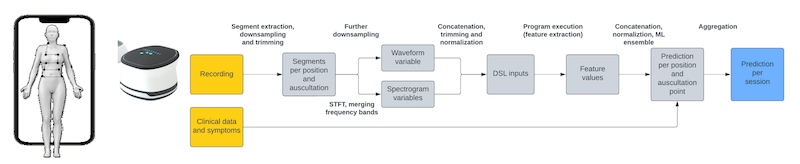
The global COVID-19 pandemic has demonstrated the urgent need for diagnostic tools that can be both readily applied and dynamically calibrated by non-specialists, in terms of a sensitivity/specificity tradeoff that complies with relevant healthcare policies and procedures. This article describes the design and deployment of a novel machine learning algorithm, Structural Machine Learning (SML), that combines memetic grammar-guided program synthesis with self-supervised learning in order to learn effectively from small data sets while remaining relatively resistant to overfitting. SML is used to construct a signal processing pipeline for audio time-series, which then serves as the diagnostic mechanism for a wide-spectrum, infrasound-to-ultrasound e-stethoscope. In blind trials supervised by a third party, SML is shown to be superior to Deep Learning approaches in terms of the area under the ROC curve, while allowing for transparent interpretation of the decision-making process.
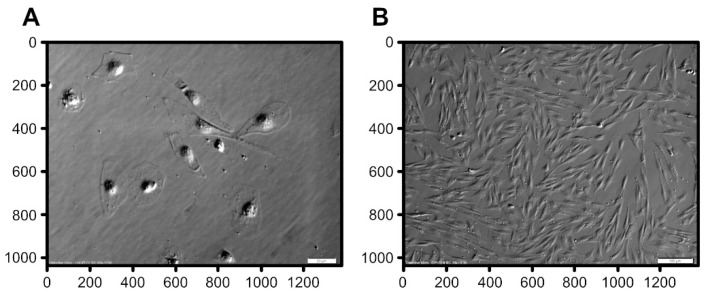
Transcriptomic and Morphological Analysis of Cells Derived from Porcine Buccal Mucosa—Studies on an In Vitro Model
Artur Bryja, Grzegorz Latosiński, Maurycy Jankowski, Ana Angelova Volponi, Paul Mozdziak, Jamil A. Shibli, Rut Bryl, Julia Spaczyńska, Hanna Piotrowska-Kempisty, Krzysztof Krawiec, Bartosz Kempisty, and Marta Dyszkiewicz-Konwińska
Animals 11.1 (2021), https://pubmed.ncbi.nlm.nih.gov/33374146/
Transcriptional analysis and live-cell imaging are a powerful tool to investigate the dynamics of complex biological systems. In vitro expanded porcine oral mucosal cells, consisting of populations of epithelial and connective lineages, are interesting and complex systems for study via microarray transcriptomic assays to analyze gene expression profile. The transcriptomic analysis included 56 ontological groups with particular focus on 7 gene ontology groups that are related to the processes of differentiation and development. Most analyzed genes were upregulated after 7 days and downregulated after 15 and 30 days of in vitro culture. The performed transcriptomic analysis was then extended to include automated analysis of differential interference contrast microscopy (DIC) images obtained during in vitro culture. The analysis of DIC imaging allowed to identify the different populations of keratinocytes and fibroblasts during seven days of in vitro culture, and it was possible to evaluate the proportion of these two populations of cells. Porcine mucosa may be a suitable model for reference research on human tissues. In addition, it can provide a reference point for research on the use of cells, scaffolds, or tissues derived from transgenic animals for applications in human tissues reconstruction.